View of Multiple Unfolded Protein Tunnels by Aggregation
Abstract
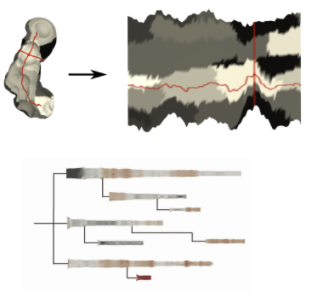
In protein dynamics, it is important to explore the accessibility of the reaction site of the protein over time. Reaction site inside the protein is usually approachable by the protein tunnel. In the paper is described a 2D representation of the tunnel, which is a result of the parametrization process of 3D mesh representation into 2 parameters (first parameter samples tunnel along the centerline, the second samples tunnel around it). The unfolded representation conveys the shape and content information of the protein tunnel surface. However, the proteins are highly flexible structures and tunnel evolves over time. One molecular dynamic simulation of the protein behaviour can have thousands of snapshots of the one tunnel. Unfortunately, the view which would convey this information is missing. As, each tunnel, can be represented by two parameters, it is possible to visualize tunnel as a tabular grid, and to do statistics visualization on each cell, communicating different properties, for example, standard deviation, min, max etc.
Context
This project is part of an ongoing research collaboration between the Masaryk University in Brno, Czech Republic. This thesis project is a follow-up to the research described in a paper [1].
Assignment
In this project, you will be working on finding new ways of visualizing the shape and content of multiple protein tunnels. You will program 2D heatmap visualizing different properties of the tunnel over time, like standard deviation of hydrophobicity in specific coordinates. The project can be extended to change the acquisition of the first tunnel parameter to be non-linear or to implement generation of average tunnel shape and encoding the information on its surface.
Requirements
Good programming skills and familiarity with C# and Unity3D are an advantage. No biology background is required.
References
- [1] Kolesar et al. “Unfolding and Interactive Exploration of Protein Tunnels and their Dynamics”, In VCBM, pp 1-10, 2016, http://www.ii.uib.no/vis/publications/publication/2016/Kolesar-2016-VCBM
Suitability
- INF219 & INF319
Contact
For more information please contact Jan Byška (jan.byska@gmail.com).
