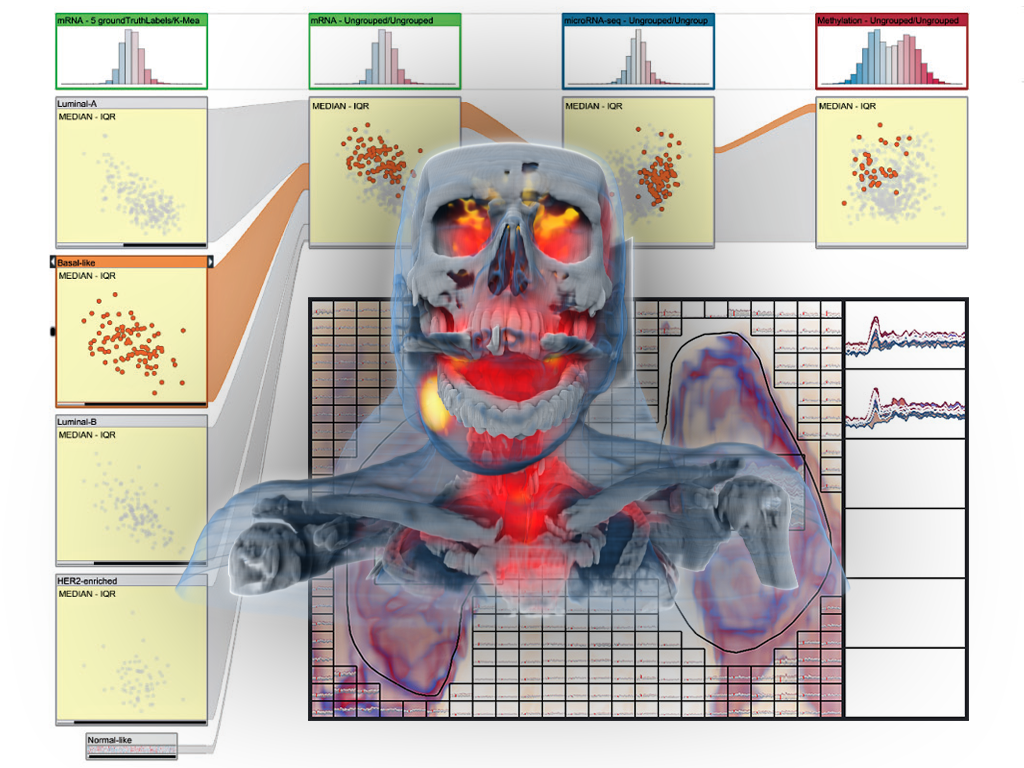
Project Overview
Technology is revolutionizing medicine. New scanners enable doctors to “look into the patient’s body” and study their anatomy and physiology without the need of a scalpel. At an amazing speed new scanning technologies emerge, providing an ever growing and increasingly varied look into medical conditions. Today, we cannot “only” look at the bones within a body, but we can also examine soft tissue, blood flow, activation networks in the brain, and many more aspects of anatomy and physiology. The increased amount and complexity of the acquired medical imaging data leads to new challenges in knowledge extraction and decision making.
In order to optimally exploit this new wealth of information, it is crucial that all this imaging data is successfully linked to the medical condition of the patient. In many cases, this is challenging, for example, when diagnosing early-stage cancer or mental disorders. Analogous to biomarkers, which are molecular structures that are used to identify medical conditions, imaging biomarkers are information structures in medical images that can help with diagnostics and treatment planning, formulated in terms of features that can be computed from the imaging data. Imaging biomarker discovery is a highly challenging task and traditionally only a single hypothesis (for a new biomarker) is examined at a time.
This makes it impossible to explore a large number as well as more complex imaging biomarkers across multi-aspect data. In the VIDI project, we propose to research and advance visual data science to improve imaging biomarker discovery through the visual integration of multi-aspect medical data with a new visualization-enabled hypothesis management framework.
We aim to reduce the time it takes to discover new imaging biomarkers by studying structured sets of hypotheses, to be examined at the same time, through the integration of computational approaches and interactive visual analysis techniques. Another related goal is to enable the discovery of more complex imaging biomarkers, across multiple modalities, that potentially are able to more accurately characterize diseases. This should lead to a new form of designing innovative and effective imaging protocols and to the discovery of new imaging biomarkers, improving suboptimal imaging protocols and thus also reducing scanning costs. Our project is a truly interdisciplinary research effort, bringing visualization research and imaging research together in one project, and this is perfectly suited for the novel Centre for Medical Imaging and Visualization that has been established in Bergen, Norway.
VIDI Approach
To achieve these goals we have divided the VIDI project into seven discrete workpackages (WP), which can be executed, to some extent, in parallel:
WP1: Hypothesis management
Research and design of methodologies necessary for structuring, representation, exploration, and analysis of hypothesis sets. Develop visual language for data interactions and methods for linking spatial with non-spatial data.
WP2: Data & Features
Exploration of medical image feature extraction and visualisation, definition of UX for selection and refinement of data features to extend into additional dimensions.
WP3: Hypotheses Scoring
Development of methods for interactive visual ranking and analyses of user hypotheses sets. Exploration of methods to provide user with evaluation preview of investigated hypotheses, linked to hypotheses visualisation and rankings.
WP4: Optimized Imaging
Evaluation of existing imaging protocols and development new imaging techniques. Investigation of imaging process to guard against suboptimal image acquisitions.
WP5: Integration
Integrate solutions from work packages 1-4.
WP6: Evalulation
Evaluate new hypothesis methods in context of three target applications: 1) gynecologic cancer; 2) neuroinflammation in MS; and 3) neurodegenerative disorders.
WP7: Management & Dissemination
Coordination between the involved partners, planning and reporting, and dissemination.
VIDI Project Team
PI: Helwig Hauser
Co-PIs: Stefan Bruckner and Renate Grüner, MMIV
Associated researcher: Noeska Smit
PhD students: Laura Garrison and Fourough Gharbalchi
This project is funded by the Bergen Research Foundation (BFS) and the University of Bergen.


Publications
2025
![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png)
@article{ziman2025genaixbiomedvis,
title={"It looks sexy but it's wrong." Tensions in creativity and accuracy using genAI for biomedical visualization},
author = {Ziman, Roxanne and Saharan, Shehryar and McGill, Ga\"{e}l and Garrison, Laura},
journal = {arXiv, IEEE Transactions on Visualization and Computer Graphics--in press},
year = {2025},
numpages = {11},
publisher = {arXiv},
doi = {10.48550/arXiv.2507.14494},
abstract = {We contribute an in-depth analysis of the workflows and tensions arising from generative AI (genAI) use in biomedical visualization (BioMedVis). Although genAI affords facile production of aesthetic visuals for biological and medical content, the architecture of these tools fundamentally limits the accuracy and trustworthiness of the depicted information, from imaginary (or fanciful) molecules to alien anatomy. Through 17 interviews with a diverse group of practitioners and researchers, we qualitatively analyze the concerns and values driving genAI (dis)use for the visual representation of spatially-oriented biomedical data. We find that BioMedVis experts, both in roles as developers and designers, use genAI tools at different stages of their daily workflows and hold attitudes ranging from enthusiastic adopters to skeptical avoiders of genAI. In contrasting the current use and perspectives on genAI observed in our study with predictions towards genAI in the visualization pipeline from prior work, our refocus the discussion of genAI's effects on projects in visualization in the here and now with its respective opportunities and pitfalls for future visualization research. At a time when public trust in science is in jeopardy, we are reminded to first do no harm, not just in biomedical visualization but in science communication more broadly. Our observations reaffirm the necessity of human intervention for empathetic design and assessment of accurate scientific visuals.},
pdf = {pdfs/ziman2025genaixbiomedvis.pdf},
images = {images/ziman2025itlookssexy.png},
thumbnails = {images/ziman2025itlookssexy_thumb.png},
project = {VIDI},
git = {https://osf.io/mbw86/}
}![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png)
@article{zhang2025deconstruct,
title = {Deconstructing Implicit Beliefs in Visual Data Journalism: Unstable Meanings Behind Data as Truth & Design for Insight},
author = {Zhang, Ke Er Amy and Jenkinson, Jodie and Garrison, Laura},
journal = {IEEE Transactions on Visualization and Computer Graphics--in press},
year = {2025},
numpages = {11},
eprint = {2507.12377},
archiveprefix = {arXiv},
primaryclass = {cs.HC},
abstract = {We conduct a deconstructive reading of a qualitative interview study with 17 visual data journalists from newsrooms across the globe. We borrow a deconstruction approach from literary critique to explore the instability of meaning in language and reveal implicit beliefs in words and ideas. Through our analysis we surface two sets of opposing implicit beliefs in visual data journalism: objectivity/subjectivity and humanism/mechanism. We contextualize these beliefs through a genealogical analysis, which brings deconstruction theory into practice by providing a historic backdrop for these opposing perspectives. Our analysis shows that these beliefs held within visual data journalism are not self-enclosed but rather a product of external societal forces and paradigm shifts over time. Through this work, we demonstrate how thinking with critical theories such as deconstruction and genealogy can reframe "success" in visual data storytelling and diversify visualization research outcomes. These efforts push the ways in which we as researchers produce domain knowledge to examine the sociotechnical issues of today's values towards datafication and data visualization. All supplemental materials for this work are available at osf.io/5fr48.},
doi = {10.48550/arXiv.2507.12377},
pdf = {pdfs/zhang2025deconstruct.pdf},
images = {images/zhang2025deconstruct.png},
thumbnails = {images/zhang2025deconstruct_thumb.png},
project = {VIDI},
git = {https://osf.io/5fr48/}
}![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png) [Bibtex]
[Bibtex] @inproceedings{zhang2025stories,
author = {Zhang, Ke Er Amy and Garrison, Laura},
title = {Modern snapshots in the crafting of a medical illustration},
booktitle = {Proceedings of CHI '25 Workshop "How do design stories work? Exploring narrative forms of knowledge in HCI"},
year = {2025},
numpages = {3},
pdf = {pdfs/zhang2025stories.pdf},
images = {images/zhang2025stories.png},
thumbnails = {images/zhang2025stories.png},
abstract = {The time-honored practice of medical illustration and visualization, has, like nearly all other disciplines, seen changes in its tooling and development pipeline in step with technological and societal developments. At its core, however, medical visualization remains a discipline focused on telling stories about biology and medicine. The story we tell in this work assumes a more distant vantage point to tell a story about the biomedical storytellers themselves. Our story peers over the shoulders of two medical illustrators in the middle of a project to illustrate a procedure in one of the small blood vessels around the heart, and through the medium of an online chat explores the dialogue, tensions, and goals of such projects in the digital age. We adopt the two-column format of the CHI template, as it is more reminiscent of the width of our usual messaging windows while working. The second part of our submission reflects on these tensions and modes of storytelling from an HCI and Visualization-situated perspective.},
project = {VIDI}
}2024
![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png) [Bibtex]
[Bibtex] @article{pokojna2024language,
title={The Language of Infographics: Toward Understanding Conceptual Metaphor Use in Scientific Storytelling},
author={Pokojn{\'a}, Hana and Isenberg, Tobias and Bruckner, Stefan and Kozl{\'i}kov{\'a}, Barbora and Garrison, Laura},
journal={IEEE Transactions on Visualization and Computer Graphics},
year={2024},
month={Oct},
publisher={IEEE},
abstract={We apply an approach from cognitive linguistics by mapping Conceptual Metaphor Theory (CMT) to the visualization domain to address patterns of visual conceptual metaphors that are often used in science infographics. Metaphors play an essential part in visual communication and are frequently employed to explain complex concepts. However, their use is often based on intuition, rather than following a formal process. At present, we lack tools and language for understanding and describing metaphor use in visualization to the extent where taxonomy and grammar could guide the creation of visual components, e.g., infographics. Our classification of the visual conceptual mappings within scientific representations is based on the breakdown of visual components in existing scientific infographics. We demonstrate the development of this mapping through a detailed analysis of data collected from four domains (biomedicine, climate, space, and anthropology) that represent a diverse range of visual conceptual metaphors used in the visual communication of science. This work allows us to identify patterns of visual conceptual metaphor use within the domains, resolve ambiguities about why specific conceptual metaphors are used, and develop a better overall understanding of visual metaphor use in scientific infographics. Our analysis shows that ontological and orientational conceptual metaphors are the most widely applied to translate complex scientific concepts. To support our findings we developed a visual exploratory tool based on the collected database that places the individual infographics on a spatio-temporal scale and illustrates the breakdown of visual conceptual metaphors.},
pdf = {pdfs/garrisonVIS24.pdf},
images = {images/garrisonVIS24.png},
thumbnails = {images/garrisonVIS24thumb.png},
project = {VIDI},
git={https://osf.io/8xrjm/}
}![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png)
@inproceedings{zimanVCBM2024mobaDash,
booktitle = {Eurographics Workshop on Visual Computing for Biology and Medicine},
editor = {Garrison, Laura and Jönsson, Daniel},
title = {{The MoBa Pregnancy and Child Development Dashboard: A Design Study}},
author = {Ziman, Roxanne and Budich, Beatrice and Vaudel, Marc and Garrison, Laura},
year = {2024},
month = {September},
publisher = {The Eurographics Association},
ISSN = {2070-5786},
ISBN = {978-3-03868-244-8},
DOI = {10.2312/vcbm.20241194},
abstract = {Visual analytics dashboards enable exploration of complex medical and genetic data to uncover underlying patterns and possible relationships between conditions and outcomes. In this interdisciplinary design study, we present a characterization of the domain and expert tasks for the exploratory analysis for a rare maternal disease in the context of the longitudinal Norwegian Mother, Father, and Child (MoBa) Cohort Study. We furthermore present a novel prototype dashboard, developed through an iterative design process and using the Python-based Streamlit App [TTK18] and Vega-Altair [VGH*18] visualization library, to allow domain experts (e.g., bioinformaticians, clinicians, statisticians) to explore possible correlations between women's health during pregnancy and child development outcomes. In conclusion, we reflect on several challenges and research opportunities for not only furthering this approach, but in visualization more broadly for large, complex, and sensitive patient datasets to support clinical research.},
pdf = {pdfs/zimanVCBM24.pdf},
images = {images/zimanVCBM24.png},
thumbnails = {images/zimanVCBM24thumb.png},
project = {VIDI},
git={https://osf.io/u6kdm/}
}![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png) [Bibtex]
[Bibtex] @MISC{zhang2024ManhattanWheel,
booktitle = {Eurographics Workshop on Visual Computing for Biology and Medicine (Posters)},
title = {{The Manhattan Wheel: A Radial Visualization Story for Genome-wide Association Study Data}},
author = {Zhang, Ke Er and Vaudel, Marc and Garrison, Laura A.},
year = {2024},
howpublished = {Poster presented at VCBM 2024.},
publisher = {The Eurographics Association},
abstract = {Genome-wide association studies (GWAS) are critical to identifying genetic variations associated with a particular trait or disease. It is important to cultivate an awareness of GWAS in the general public as members of this group are key participants of these studies. However, low genetic data literacy and trust in the sharing of genetic data pose challenges to learning and engaging with GWAS concepts. In this design study, we explore design strategies for the public communication of GWAS data. As part of this study, we present an interactive visual prototype that explores the use of narrative structure, linked visualizations through scrollytelling, and plain language to onboard and communicate genetic concepts to a GWAS-naive audience.},
pdf = {pdfs/KEZHANG_VCBM2024_Poster_ManhattanWheel.pdf},
images = {images/zhangManhattanWheel.png},
thumbnails = {images/zhangManhattanWheelthumb.png},
project = {VIDI},
git = {https://github.com/amykzhang/manhattan-wheel}
}![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png)
@inproceedings{correll2024bodydata,
abstract = {With changing attitudes around knowledge, medicine, art, and technology, the human body has become a source of information and, ultimately, shareable and analyzable data. Centuries of illustrations and visualizations of the body occur within particular historical, social, and political contexts. These contexts are enmeshed in different so-called data cultures: ways that data, knowledge, and information are conceptualized and collected, structured and shared. In this work, we explore how information about the body was collected as well as the circulation, impact, and persuasive force of the resulting images. We show how mindfulness of data cultural influences remain crucial for today's designers, researchers, and consumers of visualizations. We conclude with a call for the field to reflect on how visualizations are not timeless and contextless mirrors on objective data, but as much a product of our time and place as the visualizations of the past.},
author = {Correll, Michael and Garrison, Laura A.},
booktitle = {arXiv, Proc CHI24},
doi = {10.48550/arXiv.2402.05014},
publisher = {arXiv},
title = {When the Body Became Data: Historical Data Cultures and Anatomical Illustration},
year = {2024},
month = {Feb},
pdf = {pdfs/garrisonCHI24.pdf},
images = {images/garrisonCHI24.png},
thumbnails = {images/garrisonCHI24.png},
project = {VIDI}
}2023
![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png) [Bibtex]
[Bibtex] @MISC {balaka2023MoBaExplorer,
booktitle = {Eurographics Workshop on Visual Computing for Biology and Medicine (Posters)},
editor = {Garrison, Laura and Linares, Mathieu},
title = {{MoBa Explorer: Enabling the navigation of data from the Norwegian Mother, Father, and Child cohort study (MoBa)}},
author = {Balaka, Hanna and Garrison, Laura A. and Valen, Ragnhild and Vaudel, Marc},
year = {2023},
howpublished = {Poster presented at VCBM 2023.},
publisher = {The Eurographics Association},
abstract = {Studies in public health have generated large amounts of data helping researchers to better understand human diseases and improve patient care. The Norwegian Mother, Father and Child Cohort Study (MoBa) has collected information about pregnancy
and childhood to better understand this crucial time of life. However, the volume of the data and its sensitive nature make its
dissemination and examination challenging. We present a work-in-progress design study and accompanying web application,
the MoBa Explorer, which presents aggregated MoBa study data genotypes and phenotypes. Our research explores how to
serve two distinct purposes in one application: (1) allow researchers to interactively explore MoBa data to identify variables of
interest for further study and (2) provide MoBa study details to an interested general public.},
pdf = {pdfs/balaka2023MoBaExplorer.pdf},
images = {images/balaka2023MoBaExplorer.png},
thumbnails = {images/balaka2023MoBaExplorer-thumb.png},
project = {VIDI}
}
![[VID]](https://vis.uib.no/wp-content/papercite-data/images/video.png)
![[YT]](https://vis.uib.no/wp-content/papercite-data/images/youtube.png)