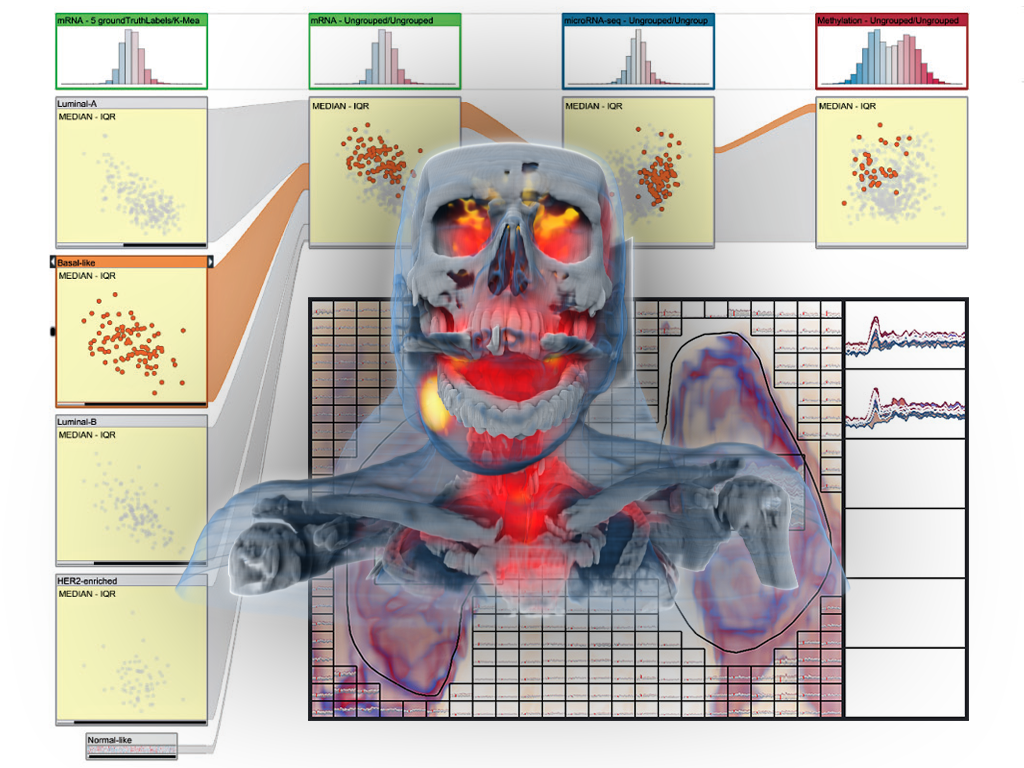
Project Overview
Technology is revolutionizing medicine. New scanners enable doctors to “look into the patient’s body” and study their anatomy and physiology without the need of a scalpel. At an amazing speed new scanning technologies emerge, providing an ever growing and increasingly varied look into medical conditions. Today, we cannot “only” look at the bones within a body, but we can also examine soft tissue, blood flow, activation networks in the brain, and many more aspects of anatomy and physiology. The increased amount and complexity of the acquired medical imaging data leads to new challenges in knowledge extraction and decision making.
In order to optimally exploit this new wealth of information, it is crucial that all this imaging data is successfully linked to the medical condition of the patient. In many cases, this is challenging, for example, when diagnosing early-stage cancer or mental disorders. Analogous to biomarkers, which are molecular structures that are used to identify medical conditions, imaging biomarkers are information structures in medical images that can help with diagnostics and treatment planning, formulated in terms of features that can be computed from the imaging data. Imaging biomarker discovery is a highly challenging task and traditionally only a single hypothesis (for a new biomarker) is examined at a time.
This makes it impossible to explore a large number as well as more complex imaging biomarkers across multi-aspect data. In the VIDI project, we propose to research and advance visual data science to improve imaging biomarker discovery through the visual integration of multi-aspect medical data with a new visualization-enabled hypothesis management framework.
We aim to reduce the time it takes to discover new imaging biomarkers by studying structured sets of hypotheses, to be examined at the same time, through the integration of computational approaches and interactive visual analysis techniques. Another related goal is to enable the discovery of more complex imaging biomarkers, across multiple modalities, that potentially are able to more accurately characterize diseases. This should lead to a new form of designing innovative and effective imaging protocols and to the discovery of new imaging biomarkers, improving suboptimal imaging protocols and thus also reducing scanning costs. Our project is a truly interdisciplinary research effort, bringing visualization research and imaging research together in one project, and this is perfectly suited for the novel Centre for Medical Imaging and Visualization that has been established in Bergen, Norway.
VIDI Approach
To achieve these goals we have divided the VIDI project into seven discrete workpackages (WP), which can be executed, to some extent, in parallel:
WP1: Hypothesis management
Research and design of methodologies necessary for structuring, representation, exploration, and analysis of hypothesis sets. Develop visual language for data interactions and methods for linking spatial with non-spatial data.
WP2: Data & Features
Exploration of medical image feature extraction and visualisation, definition of UX for selection and refinement of data features to extend into additional dimensions.
WP3: Hypotheses Scoring
Development of methods for interactive visual ranking and analyses of user hypotheses sets. Exploration of methods to provide user with evaluation preview of investigated hypotheses, linked to hypotheses visualisation and rankings.
WP4: Optimized Imaging
Evaluation of existing imaging protocols and development new imaging techniques. Investigation of imaging process to guard against suboptimal image acquisitions.
WP5: Integration
Integrate solutions from work packages 1-4.
WP6: Evalulation
Evaluate new hypothesis methods in context of three target applications: 1) gynecologic cancer; 2) neuroinflammation in MS; and 3) neurodegenerative disorders.
WP7: Management & Dissemination
Coordination between the involved partners, planning and reporting, and dissemination.
VIDI Project Team
PI: Helwig Hauser
Co-PIs: Stefan Bruckner and Renate Grüner, MMIV
Associated researcher: Noeska Smit
PhD students: Laura Garrison and Fourough Gharbalchi
This project is funded by the Bergen Research Foundation (BFS) and the University of Bergen.


Publications
2024
![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png)
@inproceedings{correll2024bodydata,
abstract = {With changing attitudes around knowledge, medicine, art, and technology, the human body has become a source of information and, ultimately, shareable and analyzable data. Centuries of illustrations and visualizations of the body occur within particular historical, social, and political contexts. These contexts are enmeshed in different so-called data cultures: ways that data, knowledge, and information are conceptualized and collected, structured and shared. In this work, we explore how information about the body was collected as well as the circulation, impact, and persuasive force of the resulting images. We show how mindfulness of data cultural influences remain crucial for today's designers, researchers, and consumers of visualizations. We conclude with a call for the field to reflect on how visualizations are not timeless and contextless mirrors on objective data, but as much a product of our time and place as the visualizations of the past.},
author = {Correll, Michael and Garrison, Laura A.},
booktitle = {arXiv, Proc CHI24},
doi = {10.48550/arXiv.2402.05014},
publisher = {arXiv},
title = {When the Body Became Data: Historical Data Cultures and Anatomical Illustration},
year = {2024},
month = {Feb},
pdf = {pdfs/garrisonCHI24.pdf},
images = {images/garrisonCHI24.png},
thumbnails = {images/garrisonCHI24.png},
project = {VIDI}
}2023
![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png) [Bibtex]
[Bibtex] @MISC {balaka2023MoBaExplorer,
booktitle = {Eurographics Workshop on Visual Computing for Biology and Medicine (Posters)},
editor = {Garrison, Laura and Linares, Mathieu},
title = {{MoBa Explorer: Enabling the navigation of data from the Norwegian Mother, Father, and Child cohort study (MoBa)}},
author = {Balaka, Hanna and Garrison, Laura A. and Valen, Ragnhild and Vaudel, Marc},
year = {2023},
howpublished = {Poster presented at VCBM 2023.},
publisher = {The Eurographics Association},
abstract = {Studies in public health have generated large amounts of data helping researchers to better understand human diseases and improve patient care. The Norwegian Mother, Father and Child Cohort Study (MoBa) has collected information about pregnancy
and childhood to better understand this crucial time of life. However, the volume of the data and its sensitive nature make its
dissemination and examination challenging. We present a work-in-progress design study and accompanying web application,
the MoBa Explorer, which presents aggregated MoBa study data genotypes and phenotypes. Our research explores how to
serve two distinct purposes in one application: (1) allow researchers to interactively explore MoBa data to identify variables of
interest for further study and (2) provide MoBa study details to an interested general public.},
pdf = {pdfs/balaka2023MoBaExplorer.pdf},
images = {images/balaka2023MoBaExplorer.png},
thumbnails = {images/balaka2023MoBaExplorer-thumb.png},
project = {VIDI}
}![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png)
@inproceedings {budich2023AIstories,
booktitle = {Eurographics Workshop on Visual Computing for Biology and Medicine},
editor = {Hansen, Christian and Procter, James and Renata G. Raidou and Jönsson, Daniel and Höllt, Thomas},
title = {{Reflections on AI-Assisted Character Design for Data-Driven Medical Stories}},
author = {Budich, Beatrice and Garrison, Laura A. and Preim, Bernhard and Meuschke, Monique},
year = {2023},
publisher = {The Eurographics Association},
ISSN = {2070-5786},
ISBN = {978-3-03868-216-5},
DOI = {10.2312/vcbm.20231216},
abstract = {Data-driven storytelling has experienced significant growth in recent years to become a common practice in various application areas, including healthcare. Within the realm of medical narratives, characters play a pivotal role in connecting audiences with data and conveying complex medical information in an engaging manner that may influence positive behavioral and lifestyle changes on the part of the viewer. However, the process of designing characters that are both informative and engaging remains a challenge. In this paper, we propose an AI-assisted pipeline for character design in the context of data-driven medical stories. Our iterative pipeline blends design sensibilities with automation to reduce the time and artistic expertise needed to develop characters reflective of the underlying data, even when that data is time-oriented as in a cohort study.},
pdf = {pdfs/budichAIstories.pdf},
images = {images/budichAIstories.png},
thumbnails = {images/budichAIstories-thumb.png},
project = {VIDI}
}![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png)
@article{mittenentzwei2023heros,
journal = {Computer Graphics Forum},
title = {{Do Disease Stories need a Hero? Effects of Human Protagonists on a Narrative Visualization about Cerebral Small Vessel Disease}},
author = {Mittenentzwei, Sarah and Weiß, Veronika and Schreiber, Stefanie and Garrison, Laura A. and Bruckner, Stefan and Pfister, Malte and Preim, Bernhard and Meuschke, Monique},
year = {2023},
publisher = {The Eurographics Association and John Wiley & Sons Ltd.},
ISSN = {1467-8659},
DOI = {10.1111/cgf.14817},
abstract = {Authors use various media formats to convey disease information to a broad audience, from articles and videos to interviews or documentaries. These media often include human characters, such as patients or treating physicians, who are involved with the disease. While artistic media, such as hand-crafted illustrations and animations are used for health communication in many cases, our goal is to focus on data-driven visualizations. Over the last decade, narrative visualization has experienced increasing prominence, employing storytelling techniques to present data in an understandable way. Similar to classic storytelling formats, narrative medical visualizations may also take a human character-centered design approach. However, the impact of this form of data communication on the user is largely unexplored. This study investigates the protagonist's influence on user experience in terms of engagement, identification, self-referencing, emotional response, perceived credibility, and time spent in the story. Our experimental setup utilizes a character-driven story structure for disease stories derived from Joseph Campbell's Hero's Journey. Using this structure, we generated three conditions for a cerebral small vessel disease story that vary by their protagonist: (1) a patient, (2) a physician, and (3) a base condition with no human protagonist. These story variants formed the basis for our hypotheses on the effect of a human protagonist in disease stories, which we evaluated in an online study with 30 participants. Our findings indicate that a human protagonist exerts various influences on the story perception and that these also vary depending on the type of protagonist.},
pdf = {pdfs/garrison-diseasestories.pdf},
images = {images/garrison-diseasestories.png},
thumbnails = {images/garrison-diseasestories-thumb.png},
project = {VIDI}
}![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png) [Bibtex]
[Bibtex] @incollection{garrison2023narrativemedvisbook,
title = {Current Approaches in Narrative Medical Visualization},
author = {Garrison, Laura Ann and Meuschke, Monique and Preim, Bernhard and Bruckner, Stefan},
year = 2023,
booktitle = {Approaches for Science Illustration and Communication},
publisher = {Springer},
address = {Gewerbestrasse 11, 6330 Cham, Switzerland},
pages = {},
note = {in publication},
editor = {Mark Roughley},
chapter = 4,
pdf = {pdfs/garrison2023narrativemedvisbook.pdf},
images = {images/garrison2023narrativemedvisbook.png},
thumbnails = {images/garrison2023narrativemedvisbook-thumb.png},
project = {VIDI}
}![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png)
@article{mittenentzwei2023investigating,
title={Investigating user behavior in slideshows and scrollytelling as narrative genres in medical visualization},
author={Mittenentzwei, Sarah and Garrison, Laura A and M{\"o}rth, Eric and Lawonn, Kai and Bruckner, Stefan and Preim, Bernhard and Meuschke, Monique},
journal={Computers \& Graphics},
year={2023},
publisher={Elsevier},
abstract={In this study, we explore the impact of genre and navigation on user comprehension, preferences, and behaviors when experiencing data-driven disease stories. Our between-subject study (n=85) evaluated these aspects in-the-wild, with results pointing towards some general design considerations to keep in mind when authoring data-driven disease stories. Combining storytelling with interactive new media techniques, narrative medical visualization is a promising approach to communicating topics in medicine to a general audience in an accessible manner. For patients, visual storytelling may help them to better understand medical procedures and treatment options for more informed decision-making, boost their confidence and alleviate anxiety, and promote stronger personal health advocacy. Narrative medical visualization provides the building blocks for producing data-driven disease stories, which may be presented in several visual styles. These different styles correspond to different narrative genres, e.g., a Slideshow. Narrative genres can employ different navigational approaches. For instance, a Slideshow may rely on click interactions to advance through a story, while Scrollytelling typically uses vertical scrolling for navigation. While a common goal of a narrative medical visualization is to encourage a particular behavior, e.g., quitting smoking, it is unclear to what extent the choice of genre influences subsequent user behavior. Our study opens a new research direction into choice of narrative genre on user preferences and behavior in data-driven disease stories.},
pdf = {pdfs/mittenentzwei2023userbehavior.pdf},
images = {images/mittenentzwei2023userbehavior.png},
thumbnails = {images/mittenentzwei2023userbehavior-thumb.png},
project = {VIDI},
doi={10.1016/j.cag.2023.06.011}
}![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png)
@article{garrison2023molaesthetics,
author={Garrison, Laura A. and Goodsell, David S. and Bruckner, Stefan},
journal={IEEE Computer Graphics and Applications},
title={Changing Aesthetics in Biomolecular Graphics},
year={2023},
volume={43},
number={3},
pages={94-101},
doi={10.1109/MCG.2023.3250680},
abstract={Aesthetics for the visualization of biomolecular structures have evolved over the years according to technological advances, user needs, and modes of dissemination. In this article, we explore the goals, challenges, and solutions that have shaped the current landscape of biomolecular imagery from the overlapping perspectives of computer science, structural biology, and biomedical illustration. We discuss changing approaches to rendering, color, human–computer interface, and narrative in the development and presentation of biomolecular graphics. With this historical perspective on the evolving styles and trends in each of these areas, we identify opportunities and challenges for future aesthetics in biomolecular graphics that encourage continued collaboration from multiple intersecting fields.},
pdf = {pdfs/garrison-aestheticsmol.pdf},
images = {images/garrison-aestheticsmol.png},
thumbnails = {images/garrison-aestheticsmol-thumb.png},
project = {VIDI}
}2022
![[PDF]](https://vis.uib.no/wp-content/plugins/papercite/img/pdf.png) [Bibtex]
[Bibtex] @phdthesis{garrison2022thesis,
title = {
From Molecules to the Masses: Visual Exploration, Analysis, and Communication
of Human Physiology
},
author = {Laura Ann Garrison},
year = 2022,
month = {September},
isbn = 9788230841389,
url = {https://hdl.handle.net/11250/3015990},
school = {Department of Informatics, University of Bergen, Norway},
abstract = {
The overarching theme of this thesis is the cross-disciplinary application of
medical illustration and visualization techniques to address challenges in
exploring, analyzing, and communicating aspects of physiology to audiences
with differing expertise.
Describing the myriad biological processes occurring in living beings over
time, the science of physiology is complex and critical to our understanding
of how life works. It spans many spatio-temporal scales to combine and bridge
the basic sciences (biology, physics, and chemistry) to medicine. Recent
years have seen an explosion of new and finer-grained experimental and
acquisition methods to characterize these data. The volume and complexity of
these data necessitate effective visualizations to complement standard
analysis practice. Visualization approaches must carefully consider and be
adaptable to the user's main task, be it exploratory, analytical, or
communication-oriented. This thesis contributes to the areas of theory,
empirical findings, methods, applications, and research replicability in
visualizing physiology. Our contributions open with a state-of-the-art report
exploring the challenges and opportunities in visualization for physiology.
This report is motivated by the need for visualization researchers, as well
as researchers in various application domains, to have a centralized,
multiscale overview of visualization tasks and techniques. Using a
mixed-methods search approach, this is the first report of its kind to
broadly survey the space of visualization for physiology. Our approach to
organizing the literature in this report enables the lookup of topics of
interest according to spatio-temporal scale. It further subdivides works
according to any combination of three high-level visualization tasks:
exploration, analysis, and communication. This provides an easily-navigable
foundation for discussion and future research opportunities for audience- and
task-appropriate visualization for physiology. From this report, we identify
two key areas for continued research that begin narrowly and subsequently
broaden in scope: (1) exploratory analysis of multifaceted physiology data
for expert users, and (2) communication for experts and non-experts alike.
Our investigation of multifaceted physiology data takes place over two
studies. Each targets processes occurring at different spatio-temporal scales
and includes a case study with experts to assess the applicability of our
proposed method. At the molecular scale, we examine data from magnetic
resonance spectroscopy (MRS), an advanced biochemical technique used to
identify small molecules (metabolites) in living tissue that are indicative
of metabolic pathway activity. Although highly sensitive and specific, the
output of this modality is abstract and difficult to interpret. Our design
study investigating the tasks and requirements for expert exploratory
analysis of these data led to SpectraMosaic, a novel application enabling
domain researchers to analyze any permutation of metabolites in ratio form
for an entire cohort, or by sample region, individual, acquisition date, or
brain activity status at the time of acquisition. A second approach considers
the exploratory analysis of multidimensional physiological data at the
opposite end of the spatio-temporal scale: population. An effective
exploratory data analysis workflow critically must identify interesting
patterns and relationships, which becomes increasingly difficult as data
dimensionality increases. Although this can be partially addressed with
existing dimensionality reduction techniques, the nature of these techniques
means that subtle patterns may be lost in the process. In this approach, we
describe DimLift, an iterative dimensionality reduction technique enabling
user identification of interesting patterns and relationships that may lie
subtly within a dataset through dimensional bundles. Key to this method is
the user's ability to steer the dimensionality reduction technique to follow
their own lines of inquiry.
Our third question considers the crafting of visualizations for communication
to audiences with different levels of expertise. It is natural to expect that
experts in a topic may have different preferences and criteria to evaluate a
visual communication relative to a non-expert audience. This impacts the
success of an image in communicating a given scenario. Drawing from diverse
techniques in biomedical illustration and visualization, we conducted an
exploratory study of the criteria that audiences use when evaluating a
biomedical process visualization targeted for communication. From this study,
we identify opportunities for further convergence of biomedical illustration
and visualization techniques for more targeted visual communication design.
One opportunity that we discuss in greater depth is the development of
semantically-consistent guidelines for the coloring of molecular scenes. The
intent of such guidelines is to elevate the scientific literacy of non-expert
audiences in the context of molecular visualization, which is particularly
relevant to public health communication.
All application code and empirical findings are open-sourced and available
for reuse by the scientific community and public. The methods and findings
presented in this thesis contribute to a foundation of cross-disciplinary
biomedical illustration and visualization research, opening several
opportunities for continued work in visualization for physiology.
},
pdf = {pdfs/garrison-phdthesis.pdf},
images = {images/garrison-thesis.png},
thumbnails = {images/garrison-thesis-thumb.png},
project = {VIDI}
}
![[VID]](https://vis.uib.no/wp-content/papercite-data/images/video.png)
![[YT]](https://vis.uib.no/wp-content/papercite-data/images/youtube.png)